

When PyMOL was still in preliminary development at version 0.99 I spent one intense week porting all the class material to PyMOL.
#INTRODUCTION TO BIOEDIT AND SWISS PDB VIEWER SOFTWARE#
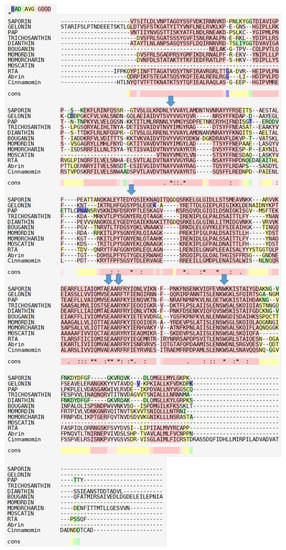
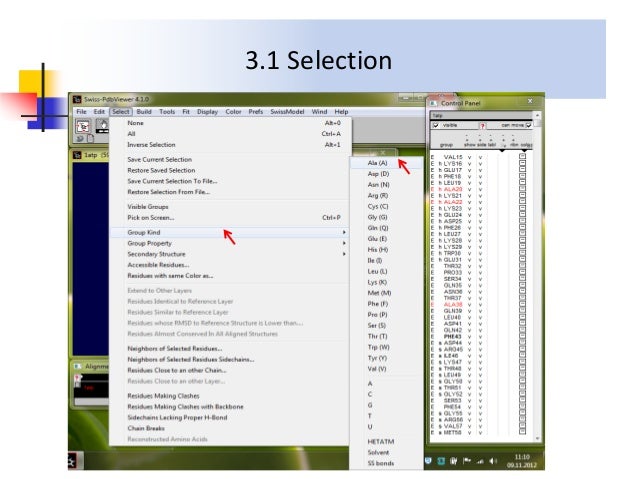
This was remedied by adding two other software in the course, one for the “publication quality renderings” ( VMD) and the other for the modeling abilities of side-chain mutations and automated 3D superimposition of structures (Swiss PDB viewer later called DeepView.) Rasmol included a sophisticated line command language but lacked the beautiful “photorealistic” renderings of the workstations.
For teaching molecular graphics on the Desktop, I first used Rasmol (see tutorial link below,) which was an amazing software fitting in about half of a 3.5” floppy disk or about 500 kilobytes. I personally used what was called “Unix Workstations” ( Silicon Graphics, sgi) which had powerful graphics and rather beautiful “ photorealistic” renderings. This class was a useful complement to the main topic of S equence Analysis and (molecular) Evolution also using computers. Ann Palmenberg classes, in particular teaching about molecular graphics on the Desktop at a time when computer use in the classroom was not yet preeminent.
The Biochem 660 PyMOL tutorial book has been split in 4 PDFs for easier read and download:


 0 kommentar(er)
0 kommentar(er)
